c-Myc Target Genes Involved in Cell Growth, Apoptosis, and Metabolism
stabilizers of MYC:Glucose, glutamine
ÇàÝïËؽµ½âc-MYC Artesunate Degrades c-MYC
chronic stress-induced cancer stem-like phenotype could be reversed by vitamin C
inducers of c-Myc:
¡¡
¡°Alternatively, combining metformin with c-MYC inhibition, prevented or reversed, respectively, resistance to metformin by enforcing their dependence on OXPHOS, suggesting a new multimodal approach for targeting the distinct metabolic features of pancreatic CSCs.¡±

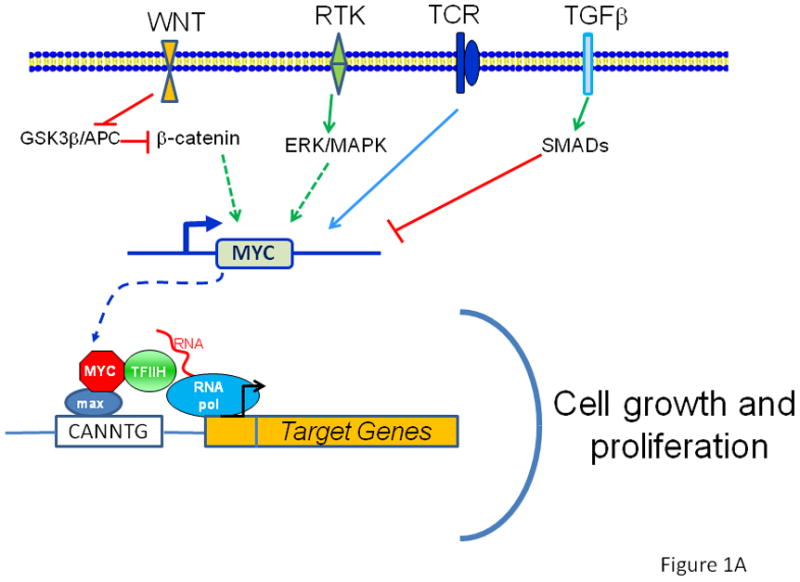
Figure 1 A. The MYC protooncogene is depicted downstream of receptor signal transduction pathways, which elicit positive or negative regulation of the MYC gene. MYC produces the transcription factor Myc, which dimerizes with Max and bind target DNA sequences or E-boxes (with the sequence 5¡ä-CANNTG-3¡ä) to regulate transcription of genes involved in cell growth and proliferation. The WNT pathway is depicted with APC negatively regulating ¦Â-catenin, which upon nuclear translocation participates in the transactivation of MYC, such that loss of APC results in constitutive oncogenic MYC expression. B. When MYC is deregulated, by gene amplication, chromosomal translocation or loss of upstream regulators, such as APC, acute sustained oncogenic MYC expression results in checkpoint activation p53 or Arf. Loss of checkpoint regulation through mutations of p53 or Arf, for example, uncloaks MYC¡¯s full tumorigenic potential.
MYC on the Path to Cancer https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3345192/
¡¡
c-Myc Target Genes Involved in Cell Growth, Apoptosis, and
Metabolism
Chi V. Dang*
The c-myc gene was discovered as the cellular homolog of the retroviral v-myc
oncogene 20 years ago (23, 25, 167). The c-myc proto-oncogene was subsequently
found to be activated in various animal and human tumors (37, 39, 42). It
belongs to the family of myc genes that includes B-myc, L-myc, N-myc, and s-myc;
however, only c-myc, L-myc, and N-myc have neoplastic potential (54, 82, 102,
118, 178). Targeted homozygous deletion of the murine c-myc gene results in
embryonic lethality, suggesting that it is critical for development (43).
Homozygous inactivation of c-myc in rat fibroblasts caused a marked prolongation
of cell doubling time, further suggesting a central role for c-myc in regulating
cell proliferation (121).
The frequency of genetic alterations of c-myc in human cancers (42) has allowed
an estimation that approximately 70,000 U.S. cancer deaths per year are
associated with changes in the c-myc gene or its expression. Given that c-myc
may contribute to one-seventh of U.S. cancer deaths, recent efforts have been
directed toward understanding the function of the c-Myc protein in cancer
biology with the hope that therapeutic insights will emerge. Past efforts, which
have contributed significantly to our current understanding of c-myc, are
discussed in a number of excellent reviews (23, 29, 37, 40, 44, 52, 66, 82, 94,
102, 118, 125, 132, 145, 178, 182, 186).
c-Myc Target Genes Involved in Cell Growth, Apoptosis, and Metabolism
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC83860/
¡¡
MYC in Regulating Immunity: Metabolism and Beyond
by J.N. Rashida Gnanaprakasam and Ruoning Wang *
Center for Childhood Cancer & Blood Diseases, Hematology/Oncology & BMT, The
Research Institute at Nationwide Children¡¯s Hospital, Ohio State University,
Columbus, OH 43205, USA
¡¡
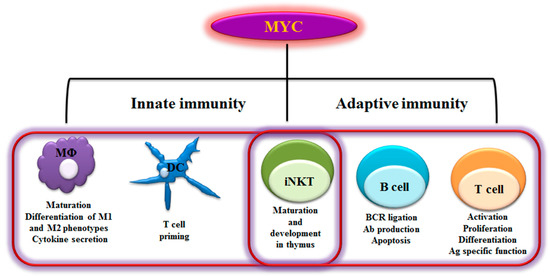
Figure 1. Myelocytomatosis oncogene (MYC)-dependent Immunity. As schematically
shown here, MYC plays a role in regulating a range of innate and adaptive immune
cells. MYC is a key transcription factor that regulates immune cell maturation,
development, proliferation and activation. M¦µ- Macrophages; DC- Dendritic cells;
iNKT- invariant Natural killer T cells; BCR- B cell receptor; Ab- Antibody; Ag-
Antigen; M1- Classically activated macrophages; M2- alternatively activated
macrophages-
Abstract: Myelocytomatosis oncogene (MYC) family members, including cellular MYC
(c-Myc), neuroblastoma derived MYC (MYCN), and lung carcinoma derived MYC
(MYCL), have all been implicated as key oncogenic drivers in a broad range of
human cancers. Beyond cancer, MYC plays an important role in other physiological
and pathological processes, namely immunity and immunological diseases. MYC
largely functions as a transcription factor that promotes the expression of
numerous target genes to coordinate death, proliferation, and metabolism at the
cellular, tissue, and organismal levels. It has been shown that the expression
of MYC family members is tightly regulated in immune cells during development or
upon immune stimulations. Emerging evidence suggests that MYC family members
play essential roles in regulating the development, differentiation and
activation of immune cells. Through driving the expression of a broad range of
metabolic genes in immune cells, MYC family members coordinate metabolic
programs to support immune functions. Here, we discuss our understanding of MYC
biology in immune system and how modulation of MYC impacts immune metabolism and
responses.
Keywords: MYC; immunity; metabolism
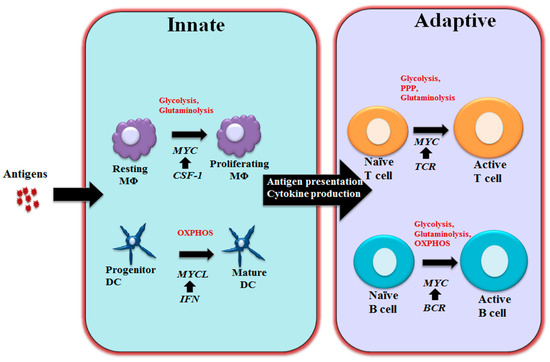
Figure 2. MYC-dependent metabolic reprograming in immunity As schematically shown here, MYC regulates immune cell metabolic reprogramming in different contexts. Colony-stimulating factor-1 (CSF-1) induces MYC expression and engages glycolysis and glutaminolysis that are required for driving macrophage proliferation. Maturation of DCs relies oxidative phosphorylation (OXPHOS) and is associated with Interferon (IFN) mediated MYCL upregulation. MYC is a component of the T cell receptor (TCR) mediated transcriptional network in T cells and coordinately controls glycolysis, pentose phosphate pathway (PPP) and glutaminolysis to support proliferation and differentiation following T cell activation. Upon BCR stimulation, upregulated MYC drives glycolysis, glutaminolysis and OXPHOS to support B cell growth and proliferation. M¦µ: Macrophages; DC: Dendritic cells; BCR: B cell receptor.
Genes | Free Full-Text | MYC in Regulating Immunity: Metabolism and Beyond |
HTML
https://www.mdpi.com/2073-4425/8/3/88/htm
Published: 27 June 2016
Glucose- and glutamine-fueled stabilization of C-Myc is required for T-cell
proliferation and malignant transformation
Dynamic regulation of cellular metabolism in response to stimulation or changes
in the environment has a crucial role in many biological processes. In
particular, T cells undergo dramatic changes in their metabolism when
progressing from quiescence to proliferation and acquisition of effector
function. This is associated with augmented flux of glucose and glutamine into
cells, and is of fundamental importance to meet the increased energy demands
associated with growth, proliferation, migration and effector molecule
production. Importantly, a similar metabolic switch occurs in cancer cells
during malignant transformation. In a study published recently in Nature
Immunology, Cantrell et al. uncover another pathway that is tightly dependent on
nutrient availability and critical for T cells.1 The authors show that increased
and sustained import of glucose and glutamine into activated T cells is
indispensable for O-GlcNAcylation, the enzymatically-mediated transfer of
N-acetylglucosamine (GlcNAc) to proteins. Multiple intracellular proteins were
found to be O-GlcNAcylated in response to T-cell receptor (TCR) signaling,
including c-Myc, an important positive regulator of cellular metabolism and
proliferation.1 O-GlcNAcylation resulted in stabilization of c-Myc, and thus
revealed an unappreciated positive-feedback loop between maintenance of cellular
metabolism and proliferation that is required for T-cell development, clonal
expansion and malignant transformation (Figure 1).
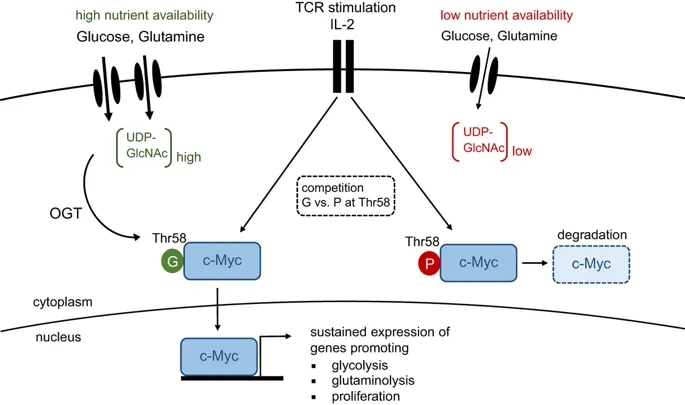
TCR activation and IL-2 signaling results in c-Myc expression, which coordinates the metabolic switch from oxidative phosphorylation to aerobic glycolysis. This process is accompanied by an increased flux of glucose and glutamine into the cell, resulting in elevated levels of UDP-GlcNAc, the end product of the hexamine biosynthetic pathway (HBP). O-GlcNAc transferase (OGT) covalently links GlcNAc sugars to proteins. Under nutrient-rich conditions, O-GlcNAcylation of Thr-58 in c-Myc outcompetes phosphorylation at the same residue, thereby preventing its degradation. Stable c-Myc expression is required for maintenance of aerobic glycolysis and proliferation upon T-cell activation. G, O-GlcNAcylation; P, phosphorylation.
In summary, the study by Cantrell et al. sheds light on a previously
unappreciated role of glycolytic metabolism in T cells. Besides being critical
for the anabolic metabolism of proliferating T cells and their effector
functions, glycolysis is also required for stabilizing c-Myc, which itself is a
critical regulator of aerobic glycolysis. Thus, O-GlcNAcylation generates a
positive-feedback loop that efficiently sustains cellular metabolism and
proliferation (Figure 1). These findings add an additional layer of complexity
to the signaling network that controls metabolic adaptation upon immune
activation and guides the differentiation of effector T cells.
Glucose- and glutamine-fueled stabilization of C-Myc is required for T-cell
proliferation and malignant transformation | Cell Death Discovery
https://www.nature.com/articles/cddiscovery201647
¡¡
O-GlcNAcylation and Metabolic Reprograming in Cancer.
Although cancer metabolism has received considerable attention over the past decade, our knowledge on its specifics is still fragmentary. Altered cellular metabolism is one of the most important hallmarks of cancer. Cancer cells exhibit aberrant glucose metabolism characterized by aerobic glycolysis, a phenomenon known as Warburg effect. Accelerated glucose uptake and glycolysis are main characteristics of cancer cells that allow them for intensive growth and proliferation. Accumulating evidence suggests that O-GlcNAc transferase (OGT), an enzyme responsible for modification of proteins with N-acetylglucosamine, may act as a nutrient sensor that links hexosamine biosynthesis pathway to oncogenic signaling and regulation of factors involved in glucose and lipid metabolism. Recent studies suggest that metabolic reprograming in cancer is connected to changes at the epigenetic level. O-GlcNAcylation seems to play an important role in the regulation of the epigenome in response to cellular metabolic status. Through histone modifications and assembly of gene transcription complexes, OGT can impact on expression of genes important for cellular metabolism. This paper reviews recent findings related to O-GlcNAc-dependent regulation of signaling pathways, transcription factors, enzymes, and epigenetic changes involved in metabolic reprograming of cancer.
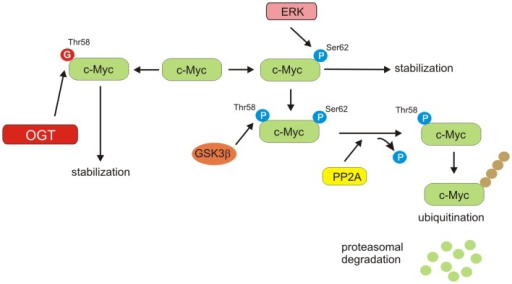
¡¡
https://openi.nlm.nih.gov/detailedresult?img=PMC4158873_fendo-05-00145-g005&req=4
¡¡
Dihydroartemisinin accelerates c-MYC oncoprotein ...
https://www.sciencedirect.com/science/article/pii/S0006295210001589
Jul 01, 2010 ¡¤ Dihydroartemisinin accelerates c-MYC oncoprotein degradation and
induces apoptosis in c-MYC-overexpressing tumor cells. ... Genes presented in
Fig. 1A are first 10 genes that are most positively correlated to the
antiproliferative activity of artesunate. The oncogene c-myc was found to be one
of the most highly positively correlated genes with ...
Cited by: 87
Publish Year: 2010
Author: Jin Jian Lu, Ling Hua Meng, Uma T. Shankavaram, Cai Hua Zhu, Lin Jiang
Tong, Guang Chen, Li Ping Lin...
(PDF) Pharmacogenomic Identification of c-Myc/Max ...
https://www.researchgate.net/publication/43352097_Pharmacogenomic_Identification_of_c...
Pharmacogenomic Identification of c-Myc/Max-Regulated Genes Associated with
Cytotoxicity of Artesunate towards Human Colon, Ovarian and Lung Cancer Cell
Lines
Artemether suppresses cell proliferation and induces ...
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5658687
We also found that ART treatment significantly decreased the expression level of
c-Myc in DLBCL. However, ART treatment did not affect the expression and the
phosphorylation of two key kinases, ERK and AKT. These results further confirmed
that c-Myc was a key downstream factor of ART in inhibiting DLBCL cell
proliferation.
Cited by: 6
Publish Year: 2017
Author: Xinying Zhao, Xudong Guo, Wenqin Yue, Jianmi
¡¡
Naturally occurring anti-cancer compounds: shining from
Chinese herbal medicine
bstract
Numerous natural products originated from Chinese herbal medicine exhibit
anti-cancer activities, including anti-proliferative, pro-apoptotic,
anti-metastatic, anti-angiogenic effects, as well as regulate autophagy, reverse
multidrug resistance, balance immunity, and enhance chemotherapy in vitro and in
vivo. To provide new insights into the critical path ahead, we systemically
reviewed the most recent advances (reported since 2011) on the key compounds
with anti-cancer effects derived from Chinese herbal medicine (curcumin,
epigallocatechin gallate, berberine, artemisinin, ginsenoside Rg3, ursolic acid,
silibinin, emodin, triptolide, cucurbitacin B, tanshinone I, oridonin, shikonin,
gambogic acid, artesunate, wogonin, ¦Â-elemene, and cepharanthine) in scientific
databases (PubMed, Web of Science, Medline, Scopus, and Clinical Trials). With a
broader perspective, we focused on their recently discovered and/or investigated
pharmacological effects, novel mechanism of action, relevant clinical studies,
and their innovative applications in combined therapy and immunomodulation. In
addition, the present review has extended to describe other promising compounds
including dihydroartemisinin, ginsenoside Rh2, compound K, cucurbitacins D, E,
I, tanshinone IIA and cryptotanshinone in view of their potentials in cancer
therapy. Up to now, the evidence about the immunomodulatory effects and clinical
trials of natural anti-cancer compounds from Chinese herbal medicine is very
limited, and further research is needed to monitor their immunoregulatory
effects and explore their mechanisms of action as modulators of immune
checkpoints.
Keywords: Cancer, Chinese herbal medicine, Natural products, Bioactive
compounds, Traditional Chinese medicine
Naturally occurring anti-cancer compounds: shining from Chinese herbal medicine
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6836491/
¡¡
¡¡
¡¡